"There are billions of neurons in our brains, but what are neurons? Just cells. The brain has no knowledge until connections are made between neurons. All that we know, all that we are, comes from the way our neurons are connected." said Tim Berners-Lee the inventor of the World Wide Web.
In order to solve difficult biological problems (e.g., creating personalized vaccines, understanding protein sequences, cancer research, etc.) with artificial intelligence (AI), machine learning algorithms must first understand the input data (inputs) that define the problem.
Neural networks (a biologically-inspired programming paradigm) and deep learning currently provide the best solutions to solve many problems in biology and medicine (e.g., genomics, population genetics, neuroscience…).
Brain: an apparatus with which we think we think
~ Ambrose Bierce, The Devil’s Dictionary
The AI model has to remember the contextual past in order to generate and predict the contextual sequences of the future. The model is more robust against mistakes when having longer memory/pattern ranges (i.e., learns from large amounts of data).
 Generating Sequences with Recurrent Neural Networks (RNN). Source:
Graves 2012
Generating Sequences with Recurrent Neural Networks (RNN). Source:
Graves 2012
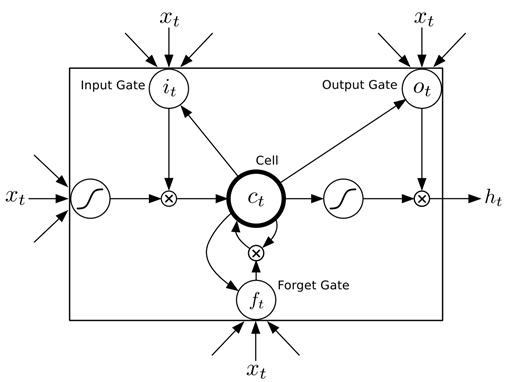
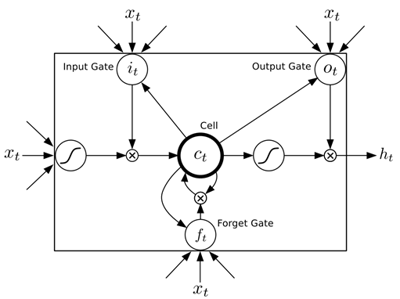
For general problem solving the Long Short-Term Memory (LSTM) model can be used. LSTM is a Recurrent Neural Network (RNN) model, designed to have a better memory. It uses memory cells surrounded by gate units to store, read, write, and reset data sequences (see figure).
One of the most commonly used programming languages in life sciences is Python. Python is an object-oriented programming language that is ideal for biological data analysis (e.g., handling nucleic acid and protein sequences, reading, parsing and handling biological sequence data files, searching even the largest biological sequences quickly and efficiently, molecular modeling, managing plate assay data…).
To quickly start problem solving using deep/machine learning algorithms and methods, an open source Keras library can be used. Keras is a minimalistic, modular, and extensive deep learning Phython library running on top of Theano and TensorFlow. It was developed with a focus on enabling fast experimentation. Keras was initially developed as part of the research effort of project ONEIROS (Open-ended Neuro-Electronic Intelligent Robot Operating System)
Key features of the library:
- Allows easy and fast prototyping (through total modularity, minimalism, and extensibility)
- Supports arbitrary connectivity schemes, including multi-input and multi-output training
- Supports both convolutional and recurrent networks, as well as combination of the two
- Runs seamlessly on CPU and GPU
- Keras is compatible with Python 2.7-3.5
The Modularity of Keras allows combining neural layers with cost functions, optimizers, activation functions, regularization schemes, etc. when creating new deep learning models.
Keras/TensorFlow/Theano has been used in different experiments (e.g., Cancer Immunology, State-of-the-art predictions in various regulatory effects directly from DNA sequences, Unsupervised feature construction and knowledge extraction from genome-wide assays of breast cancer with denoising autoencoders…).
I believe that once the machine learning methods, tools, and algorithms are more widely used by biologists and other life scientists they can considerably accelerate experimentation, research, and improve medical diagnostics.
Boris Beric, MSc
Further reading:
- Graves, Generating Sequences with Recurrent Neural Networks, 2012
- Tan, M. Ung, C. Cheng, C. S. Greene, Unsupervised feature construction and knowledge extraction from genome-wide assays of breast cancer with denoising autoencoders, Pacific Symposium on Biocomputing 2015
- Rubinsteyn, PyData NYC 2015
- Rampasek, A. Goldenberg, TensorFlow: Biology’s Gateway to Deep Learning, Cell Systems 2, January 27, 2016
- Ray Kurzweil, How to Create a Mind, Viking-Penguin Group, 2012
[tw_callout size="waves-shortcode" text="" callout_style="style2" thumb="" btn_text="Republish the article" color="#37a0d9" btn_url="https://scinote.net/blog/republish/" btn_target="_blank"]